12. dask and Large Datasets Computations#
Some people think that large datasets or “big data” are mostly applied to machine learning and artificial intelligence fields. In atmospheric science,
Big data refers to data sets that are so voluminous and complex that traditional data processing application software is inadequate to deal with them.
Due to the long periods and finer grid resolutions of modern reanalysis data or model outputs, it often leads to system overload if not processed properly. You may see the following error message:
MemoryError: Unable to allocate 52.2 GiB for an array with shape (365, 37, 721, 1440) and data type float32
This error message appears because the data size has exceeded the RAM capacity. How should we avoid this situation?
Dask#
`dask`` is a flexible library for parallel computing in Python. It can scale up to operate on large datasets and perform computations that cannot fit into memory. dask achieves this by breaking down large computations into smaller tasks, which are then executed in parallel. This can avoid consuming large amount of RAM.
To understand the usage of dask, with demonstrate first with a 1000 × 4000 array size.
1. Numpy Array:
import numpy as np
shape = (1000, 4000)
ones_np = np.ones(shape)
ones_np
array([[1., 1., 1., ..., 1., 1., 1.],
[1., 1., 1., ..., 1., 1., 1.],
[1., 1., 1., ..., 1., 1., 1.],
...,
[1., 1., 1., ..., 1., 1., 1.],
[1., 1., 1., ..., 1., 1., 1.],
[1., 1., 1., ..., 1., 1., 1.]])
2. Dask Array:
import dask.array as da
ones = da.ones(shape)
ones
|
||||||||||||||||
Dask devides the entire array into sub-arrays named “chunk”. In dask, we can specify the size of a chunk.
chunk_shape = (1000, 1000)
ones = da.ones(shape, chunks=chunk_shape)
ones
|
||||||||||||||||
We can do some arithmetic calculations, such as multiplication and averaging.
ones_mean = (ones * ones[::-1, ::-1]).mean()
ones_mean
|
||||||||||||||||
Following is the calculation procedure:
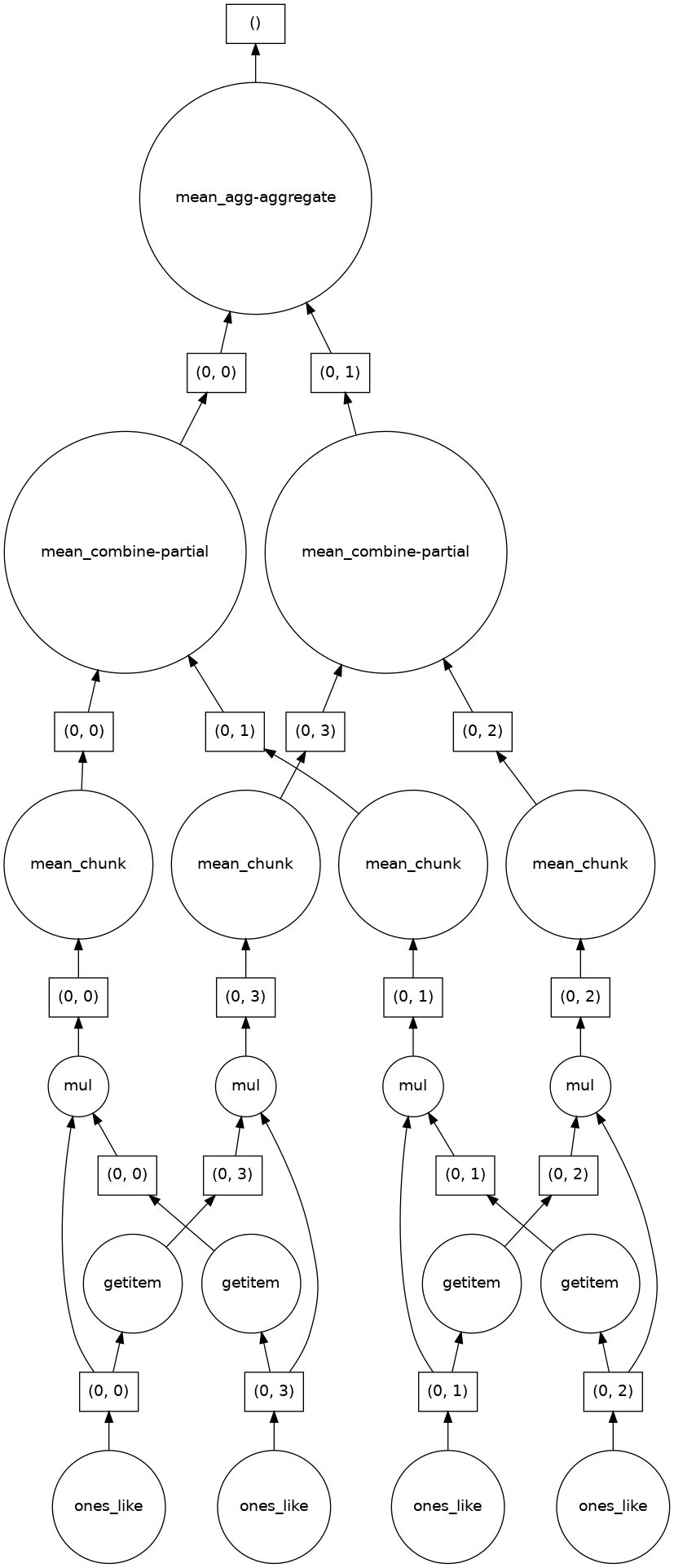
Dask allows computation of each chunk in each memory core, and finally combines all the computation of each chunk to a final result. For more advanced usages of dask, see dask official tutorial.
Dask integrates commonly-used functions in numpy and xarray, which is beneficial to processing climate data. Then how will dask help with large datasets?
Large Climate Dataset Processing#
In Unit 2, we introduced the parallel=True option in xarray.open_mfdataset. This option allows xarray to read the file using dask array. Therefore, the system will read data using multi-core computation to speed up the reading process. Now, we read the NCEP R2 850-hPa wind field and specify the size of chunks by chunks={'time': 183, 'level': 1, 'longitude': 93*2, 'latitude': 91*2} then calculate climatology, anomaly, and plot.
import xarray as xr
import dask
with dask.config.set(**{'array.slicing.split_large_chunks': False}):
uds = xr.open_mfdataset('./data/ncep_r2_uv850/u850.*.nc',
combine = "by_coords",
parallel=True,
chunks={'time':183, 'longitude':36,'latitude':24}
)
vds = xr.open_mfdataset('data/ncep_r2_uv850/v850.*.nc',
combine = "by_coords",
parallel=True,
chunks={'time':183, 'longitude':36,'latitude':24}
)
%time
CPU times: user 10 μs, sys: 2 μs, total: 12 μs
Wall time: 24.1 μs
u = uds.sel(lat=slice(90,0)).uwnd
v = vds.sel(lat=slice(90,0)).vwnd
%time
u
CPU times: user 10 μs, sys: 2 μs, total: 12 μs
Wall time: 22.6 μs
<xarray.DataArray 'uwnd' (time: 8766, level: 1, lat: 37, lon: 144)> Size: 187MB
dask.array<getitem, shape=(8766, 1, 37, 144), dtype=float32, chunksize=(183, 1, 37, 144), chunktype=numpy.ndarray>
Coordinates:
* time (time) datetime64[ns] 70kB 1998-01-01 1998-01-02 ... 2021-12-31
* lon (lon) float32 576B 0.0 2.5 5.0 7.5 10.0 ... 350.0 352.5 355.0 357.5
* lat (lat) float32 148B 90.0 87.5 85.0 82.5 80.0 ... 7.5 5.0 2.5 0.0
* level (level) float32 4B 850.0
Attributes: (12/14)
standard_name: eastward_wind
long_name: Daily U-wind on Pressure Levels
units: m/s
unpacked_valid_range: [-140. 175.]
actual_range: [-78.96 110.35]
precision: 2
... ...
var_desc: u-wind
dataset: NCEP/DOE AMIP-II Reanalysis (Reanalysis-2) Daily A...
level_desc: Pressure Levels
statistic: Mean
parent_stat: Individual Obs
cell_methods: time: mean (of 4 6-hourly values in one day)Now we see the chunk information. This is because of the dask lazy computation that the program didn’t really compute with real data values. At this moment, the data is not loaded in the RAM of the system.
Now, we are going to calculate the climatological mean. However, the built-in functions groupby() and resample() in xarray are not well supported. Therefore, we will compute with the flox libray, which is a library with significantly faster groupby calculations. The flox groupby method is flox.xarray.xarray_reduce.
from flox.xarray import xarray_reduce
uDayClm = xarray_reduce(u,u.time.dt.dayofyear,func='mean',dim='time')
vDayClm = xarray_reduce(u,u.time.dt.dayofyear,func='mean',dim='time')
uDayClm
<xarray.DataArray 'uwnd' (dayofyear: 366, level: 1, lat: 37, lon: 144)> Size: 8MB
dask.array<transpose, shape=(366, 1, 37, 144), dtype=float32, chunksize=(366, 1, 37, 144), chunktype=numpy.ndarray>
Coordinates:
* lon (lon) float32 576B 0.0 2.5 5.0 7.5 ... 350.0 352.5 355.0 357.5
* lat (lat) float32 148B 90.0 87.5 85.0 82.5 80.0 ... 7.5 5.0 2.5 0.0
* level (level) float32 4B 850.0
* dayofyear (dayofyear) int64 3kB 1 2 3 4 5 6 7 ... 361 362 363 364 365 366
Attributes: (12/14)
standard_name: eastward_wind
long_name: Daily U-wind on Pressure Levels
units: m/s
unpacked_valid_range: [-140. 175.]
actual_range: [-78.96 110.35]
precision: 2
... ...
var_desc: u-wind
dataset: NCEP/DOE AMIP-II Reanalysis (Reanalysis-2) Daily A...
level_desc: Pressure Levels
statistic: Mean
parent_stat: Individual Obs
cell_methods: time: mean (of 4 6-hourly values in one day)After lazy computation, we can now really trigger computation and return the final result with .compute().
uDayClm_fn = uDayClm.compute()
vDayClm_fn = vDayClm.compute()
uDayClm_fn
<xarray.DataArray 'uwnd' (dayofyear: 366, level: 1, lat: 37, lon: 144)> Size: 8MB
array([[[[-1.5247940e+00, -1.4514602e+00, -1.3754174e+00, ...,
-1.7235428e+00, -1.6562518e+00, -1.5960444e+00],
[-5.6333429e-01, -4.1833475e-01, -2.7458504e-01, ...,
-9.6874976e-01, -8.4125155e-01, -6.9416922e-01],
[ 7.5583190e-01, 9.2770594e-01, 1.0993727e+00, ...,
3.0166462e-01, 4.4770643e-01, 6.0145718e-01],
...,
[-1.4350017e+00, -1.3072938e+00, -1.2437533e+00, ...,
-3.2308366e+00, -2.5950010e+00, -1.8743768e+00],
[-7.3541850e-01, -6.0729337e-01, -4.0958592e-01, ...,
-2.8920867e+00, -2.0904186e+00, -1.2187523e+00],
[-6.3625211e-01, -6.9604486e-01, -6.7854387e-01, ...,
-2.7762527e+00, -1.9468769e+00, -1.0535429e+00]]],
[[[ 1.3624781e-01, 1.8458217e-01, 2.3916422e-01, ...,
-1.3961315e-02, 3.6873043e-02, 8.3957411e-02],
[ 2.3895724e-01, 3.3770636e-01, 4.3708053e-01, ...,
-5.7292778e-02, 4.5207102e-02, 1.4082973e-01],
[ 2.3937146e-01, 3.5499719e-01, 4.8520657e-01, ...,
...
[-1.4406258e+00, -1.0618763e+00, -4.6687624e-01, ...,
-3.2627106e+00, -2.4233358e+00, -1.8195858e+00],
[-8.7166828e-01, -7.2979182e-01, -4.7916898e-01, ...,
-2.8472936e+00, -1.9472933e+00, -1.2368768e+00]]],
[[[-4.1091676e+00, -3.9441681e+00, -3.7783356e+00, ...,
-4.5566688e+00, -4.4050002e+00, -4.2674980e+00],
[-3.2116699e+00, -3.1075017e+00, -2.9991691e+00, ...,
-3.5141671e+00, -3.4233353e+00, -3.3258324e+00],
[-3.0250015e+00, -3.1033335e+00, -3.1783364e+00, ...,
-2.5975020e+00, -2.7550004e+00, -2.8883379e+00],
...,
[ 6.7749852e-01, 9.3666548e-01, 8.9166456e-01, ...,
-2.1883342e+00, -1.1716664e+00, -9.6667372e-02],
[ 3.4666610e-01, 7.5583148e-01, 1.2258316e+00, ...,
-2.0166693e+00, -1.1691660e+00, -3.0000091e-01],
[ 2.3749900e-01, 4.7500062e-01, 9.8333144e-01, ...,
-1.7283325e+00, -9.7916764e-01, -2.2666772e-01]]]],
dtype=float32)
Coordinates:
* lon (lon) float32 576B 0.0 2.5 5.0 7.5 ... 350.0 352.5 355.0 357.5
* lat (lat) float32 148B 90.0 87.5 85.0 82.5 80.0 ... 7.5 5.0 2.5 0.0
* level (level) float32 4B 850.0
* dayofyear (dayofyear) int64 3kB 1 2 3 4 5 6 7 ... 361 362 363 364 365 366
Attributes: (12/14)
standard_name: eastward_wind
long_name: Daily U-wind on Pressure Levels
units: m/s
unpacked_valid_range: [-140. 175.]
actual_range: [-78.96 110.35]
precision: 2
... ...
var_desc: u-wind
dataset: NCEP/DOE AMIP-II Reanalysis (Reanalysis-2) Daily A...
level_desc: Pressure Levels
statistic: Mean
parent_stat: Individual Obs
cell_methods: time: mean (of 4 6-hourly values in one day)The computed result is now saved in a DataArray.
Best Practices with Working with dask#
Save intermediate results to disk as a netCDF files (using
to_netcdf()) and then load them again withopen_dataset()for further computations.Specify smaller chunks across space when using
open_mfdataset()(e.g.,chunks={'latitude': 10, 'longitude': 10})。Chunk as early as possible, and avoid rechunking as much as possible. Always pass the
chunks={}argument toopen_mfdataset()to avoid redundant file reads.groupby()is a costly operation and will perform a lot better if the flox package is installed. See thefloxdocumentation for more. By default Xarray will usefloxif installed.
